Exercise solution for Chapter 2, Part 2
Exercise 2.6
The first part of the exercise asks you to:
Choose your own prior for the parameters of the beta distribution. You can do this by sketching it here: https://jhubiostatistics.shinyapps.io/drawyourprior.
After sketching a plot, I chose the parameters to set up a prior:
Using this prior
Next, the exercise asks you:
Once you have set up a prior, re-analyse the data from Section 2.9.2, where we saw Y = 40 successes out of n = 300 trials.
To be able to use the loglikelihood function from the text, I first needed to redefine it here:
loglikelihood = function(theta, n = 300, k = 40) { ## Function definition from the textbook
log(choose(n, k)) + k * log(theta) + (n - k) * log(1 - theta)
}Then, I created a vector of
thetas = seq(0, 1, by = 0.001)
plot(thetas, loglikelihood(thetas), xlab = expression(theta),
ylab = expression(paste("log f(", theta, " | y)")),type = "l")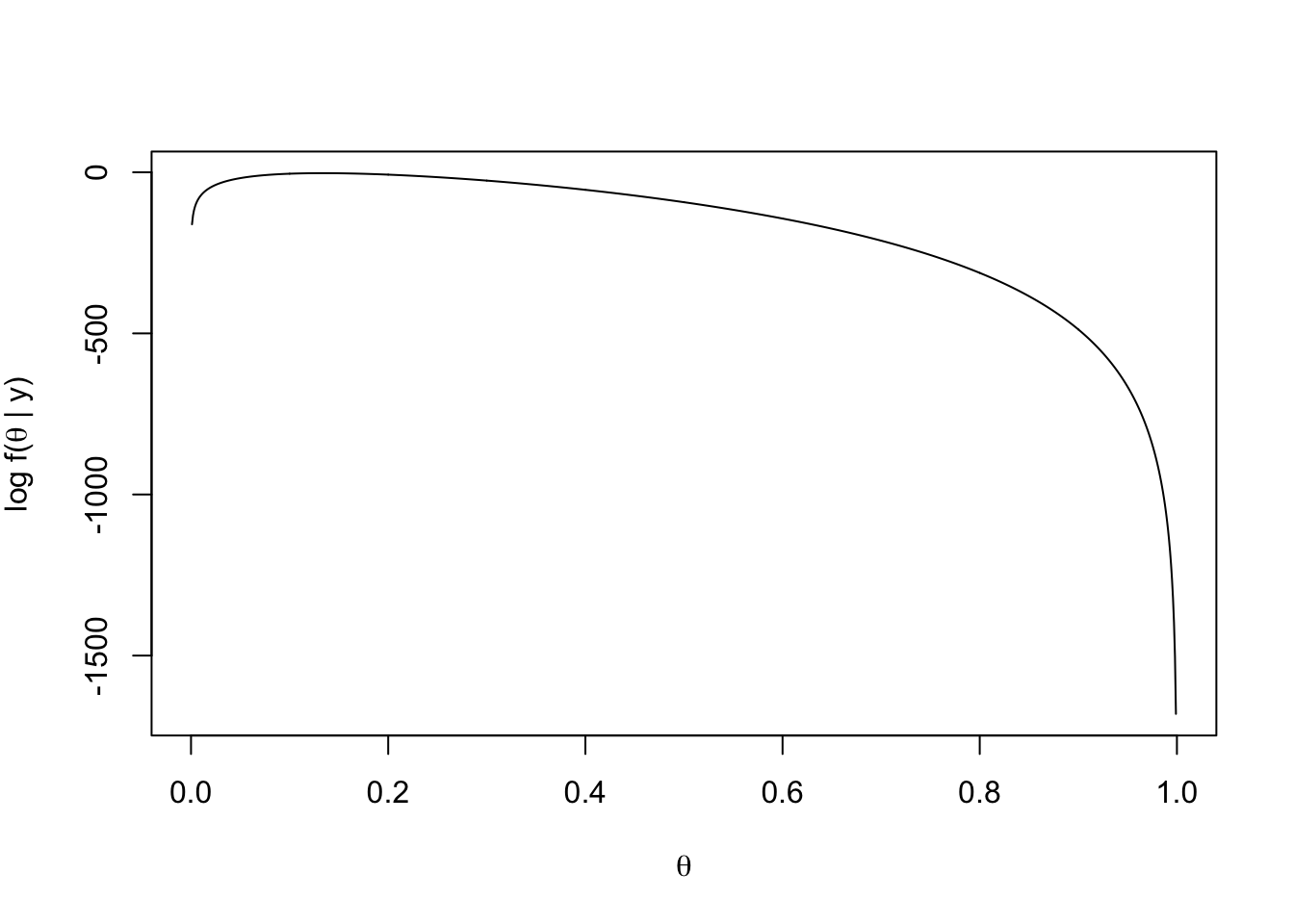
Next, I used rbeta to draw 1,000,000 random samples from a beta distribution with my new picks for the parameters for
rtheta = rbeta(1000000, shape1 = 2.47, shape2 = 8.5)After running the above, for each of these
y = vapply(rtheta, function(th) {
rbinom(1, prob = th, size = 300)
}, numeric(1))
hist(y, breaks = 50, col = "orange", main = "", xlab = "")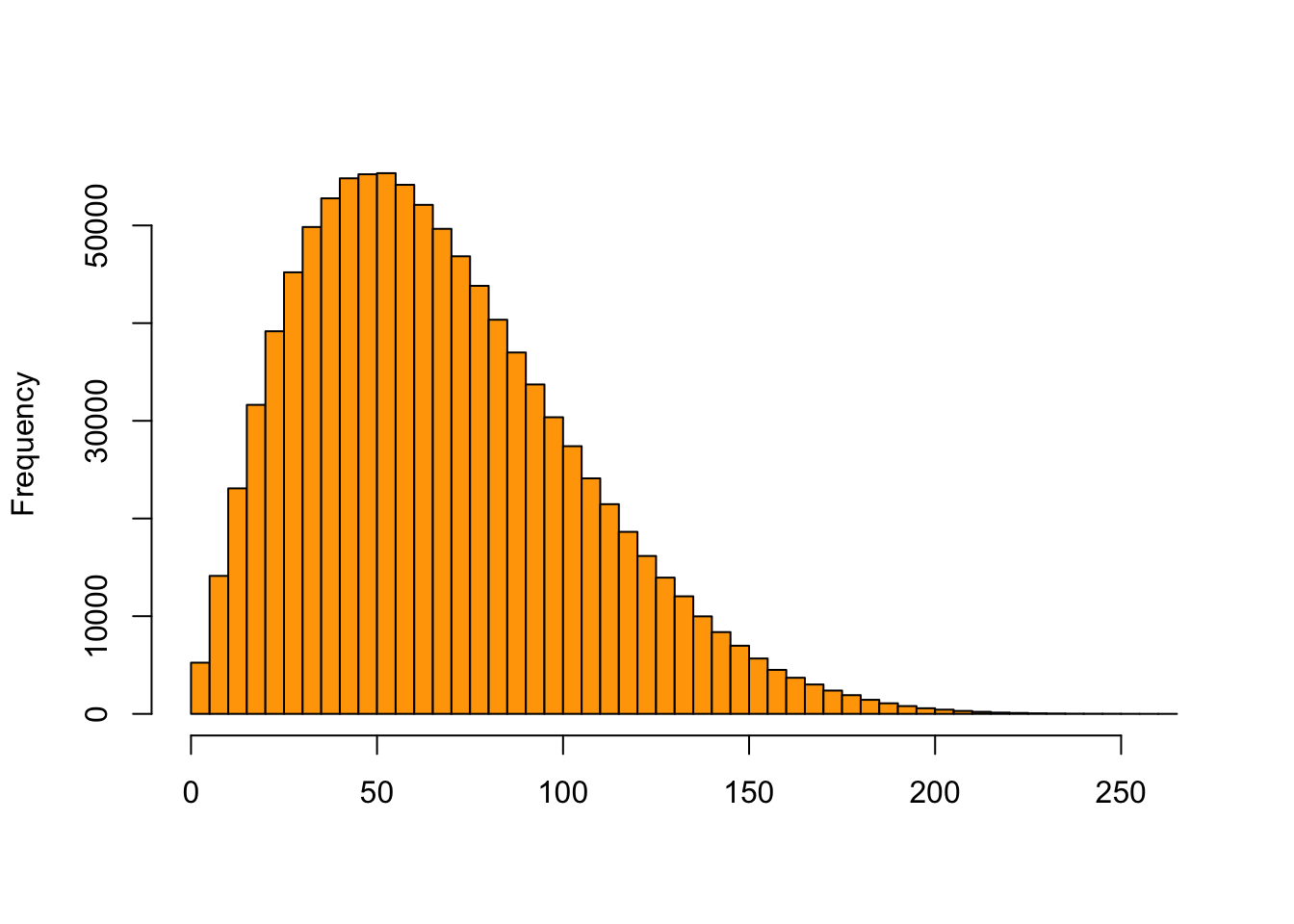
Our next step is to use this information to generate a posterior distribution of
After running the above, for each of these thetas, we generated simulated values for the posterior distribution of
thetaPostEmp = rtheta[ y == 40 ]
hist(thetaPostEmp, breaks = 40, col = "chartreuse4", main = "",
probability = TRUE, xlab = expression("posterior"~theta), ylim=c(0,40))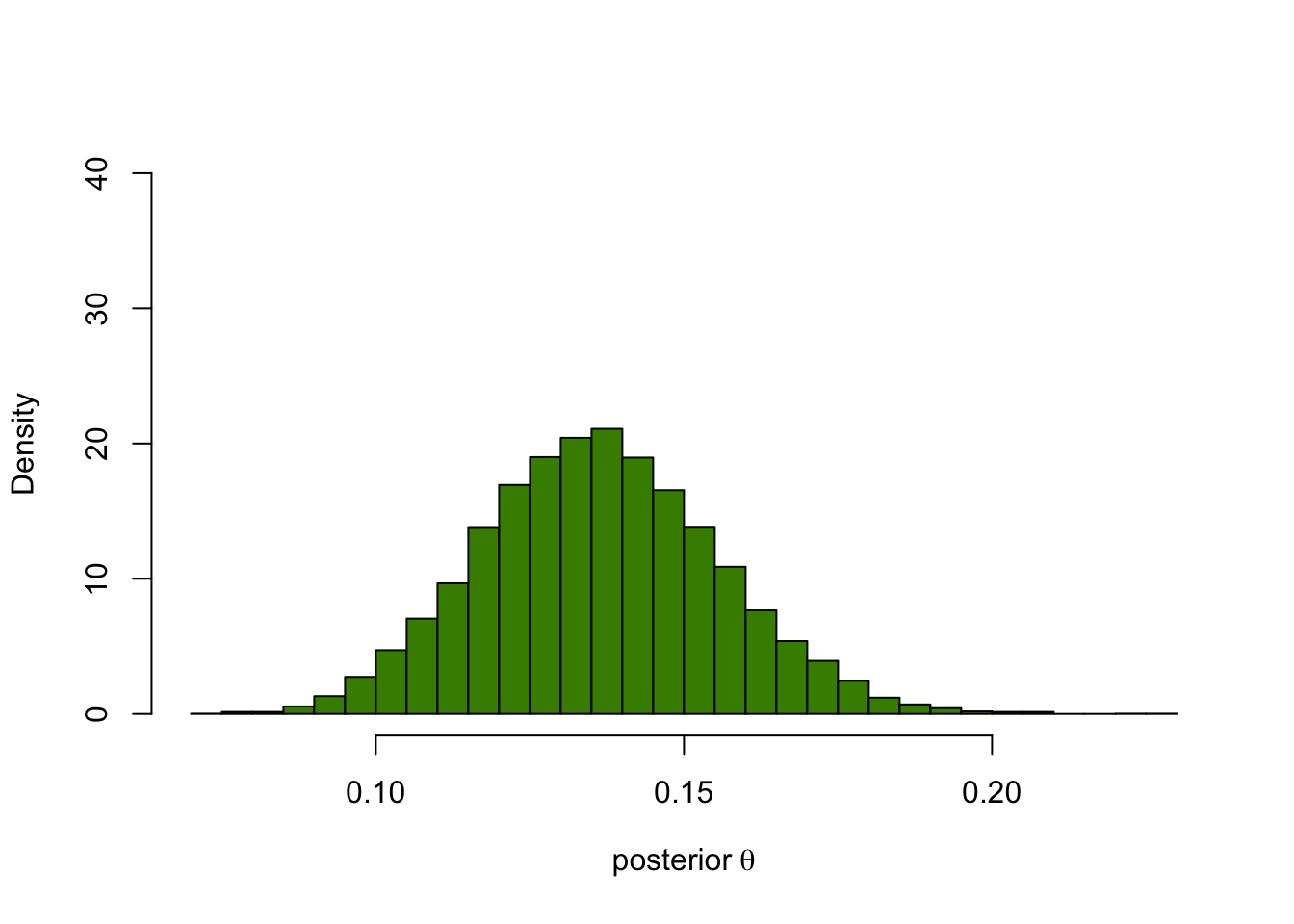
densPostTheory = dbeta(thetas, 42.47, 268.5)You can check how this compares to the theoretical posterior distribution
for
hist(thetaPostEmp, breaks = 40, col = "chartreuse4", main = "",
probability = TRUE, xlab = expression("posterior"~theta))
lines(thetas, densPostTheory, type="l", lwd = 3)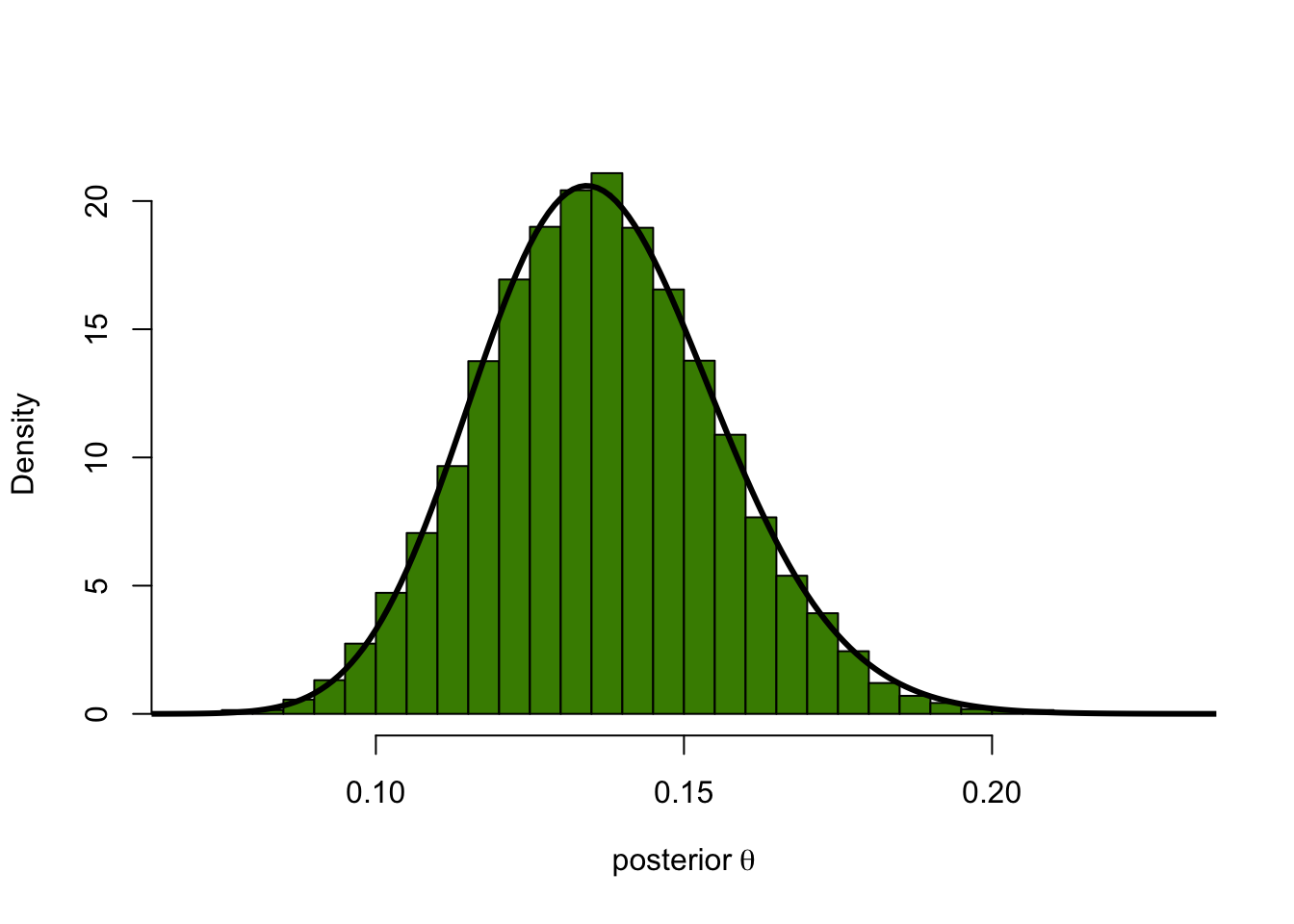
We can also check the means of both distributions computed above.
mean(thetaPostEmp) # Empirical## [1] 0.1363246dtheta = thetas[2]-thetas[1]
sum(thetas * densPostTheory * dtheta) # Theoretical## [1] 0.1365727Monte Carlo integration
We can use Monte Carlo integration instead and then check the agreement between our Monte Carlo sample thetaPostMC and our sample thetaPostEmp with a QQ plot:
thetaPostMC = rbeta(n = 1e6, 42.47, 268.5)
mean(thetaPostMC)## [1] 0.1365813qqplot(thetaPostMC, thetaPostEmp, type = "l", asp = 1)
abline(a = 0, b = 1, col = "blue")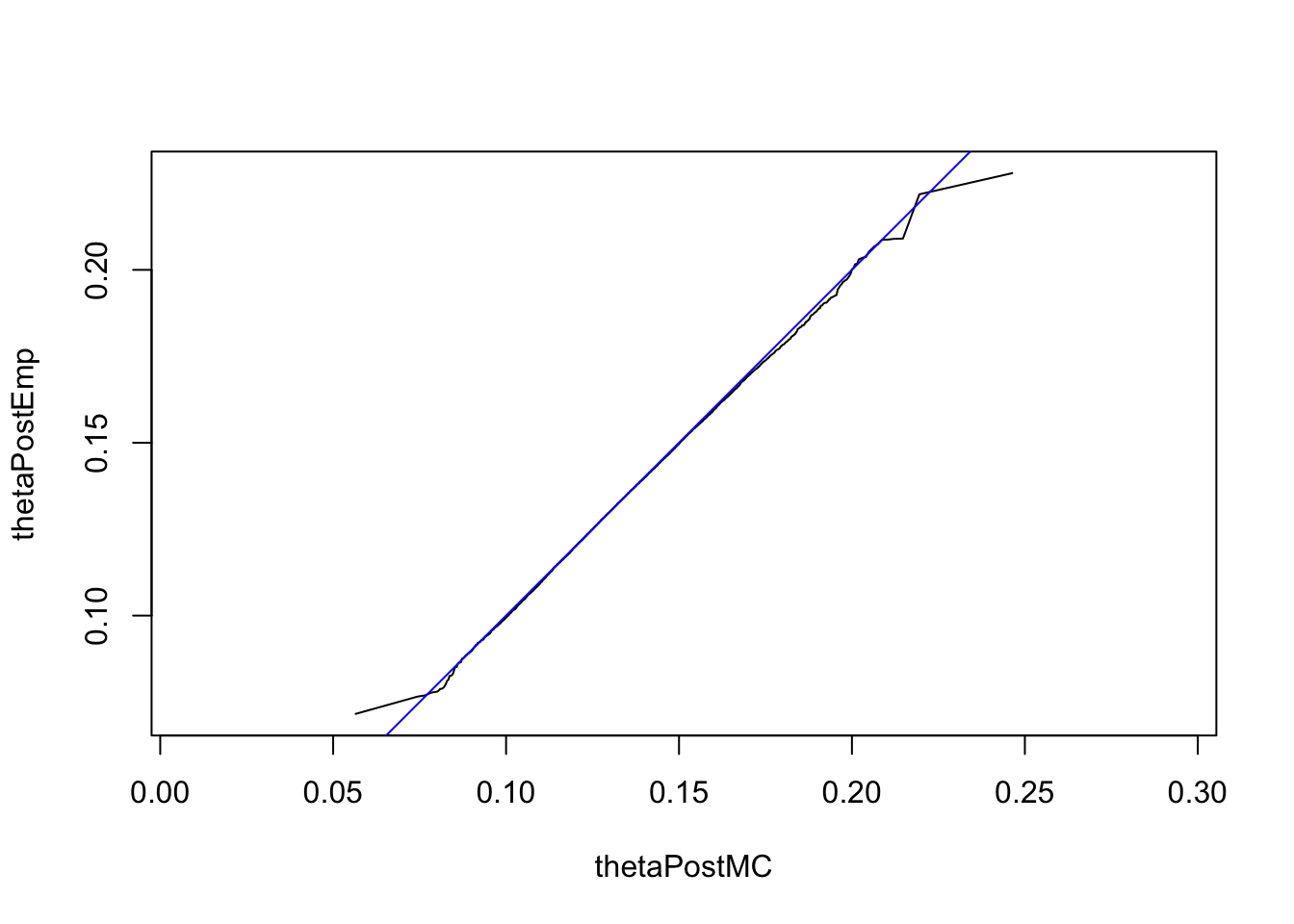
densPost2 = dbeta(thetas, 42.47, 268.5)
mcPost2 = rbeta(1e6, 42.47, 268.5)
sum(thetas * densPost2 * dtheta) # mean, by numeric integration## [1] 0.1365727mean(mcPost2) # mean, by MC## [1] 0.1365487thetas[which.max(densPost2)] # MAP estimate## [1] 0.134quantile(mcPost2, c(0.025, 0.975))## 2.5% 97.5%
## 0.1007321 0.1767921